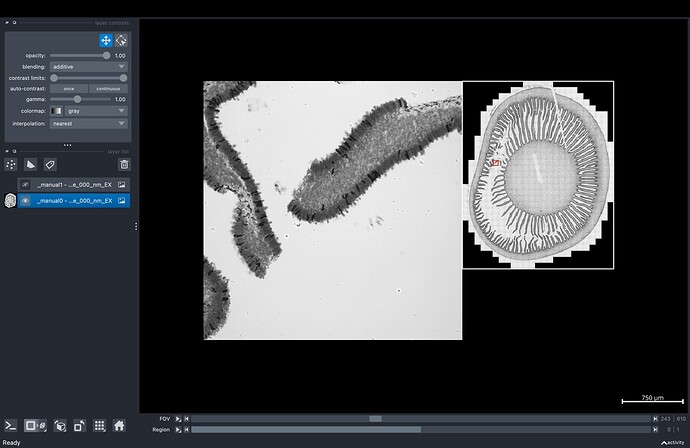
We’ve developed a navigation system for ndviewer that handles 5-D microscopy acquisitions (t,r,f,z,c,y,x). If the user decides to click on the “Downsampled Navigator” checkbox, the system creates an interactive napari layer that allows “jumping” between FOVs by clicking on it (see GUI image).
Modules
- Downsampler (downsampler.py)
- Creates 75x75 pixel tiles from full-resolution images
- Uses parallel processing with ThreadPoolExecutor
- Caches tiles to disk (cache/nav_{region}fov{fov}_{tile_size}.npy)
- Navigator Overlay (navigator.py)
- Manages multi-channel navigator layers in napari
- Handles click-to-navigate interactions
- Synchronizes contrast between FOV and navigator
- ndviewer (ndviewer_napari/fov_viewer.py)
- Lazy-loads TIFF files using dask arrays
- Creates separate layers per channel with proper colormaps
Customizable Parameters:
tile_size: Default 75 pixels (balance of speed/quality)
n_workers: Thread count (default: CPU count)
cache_enabled: Disk caching on/off
Usage:
Drop your acquisition folder onto the launcher window. Click checkbox
Click any tile in the navigator to jump to that FOV
Toggle visibility with eye icons, adjust contrast with sliders
Use the Region slider to switch between different regions
Expected File Structure
acquisition_dir/
├── 0/ # Timepoint directory
│ ├── coordinates.csv # FOV positions
│ └── {region}{fov}{z}{modality}{channel}.tiff
├── acquisition parameters.json
└── configurations.xml
Output
Navigator creates tile_cache/ directory with pre-computed tiles
No modifications to original data
All processing done on-the-fly or cached
No GPU neededx