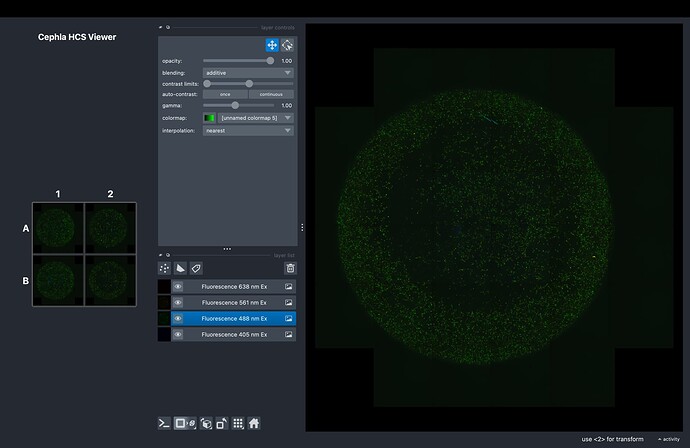
We’ve developed an HCS (High Content Screening) viewer that displays stitched OME-ZARR datasets as interactive well plate grids with real-time thumbnail synchronization. Users can click any well thumbnail to open it in an embedded napari viewer.
Usage:
- Drop dataset folder onto launcher or specify path
- Click any well thumbnail to view in napari
- Adjust contrast/visibility - changes apply to all thumbnails
- Cycle through wells
- Toggle layout orientation with right-click menu
Expected File Structure
dataset_dir/
├── 0_stitched/ # Timepoint directory
│ ├── A1.ome.zarr # Well regions
│ ├── A2.ome.zarr
│ └── …
├── 1_stitched/ # Additional timepoints
└── …
Output
- In-memory caching for fast thumbnail updates
- No modifications to original OME-ZARR data
- Real-time contrast/visibility synchronization
- Cross-platform Qt6 interface
Modules:
Grid Display (WellPlateWidget)
- Renders wells in standard plate formats (4, 6, 12, 24, 96, 384, 1536-well)
- Generates composite RGB thumbnails from multi-channel data
- Hover and click-to-open functionality
Thumbnail Generation (ThumbnailWorker)
- Loads OME-ZARR data with parallel processing (ThreadPoolExecutor)
- Caches channel data in memory for instant updates
- Applies contrast limits and visibility before compositing
Contrast Synchronization (GridViewerGUI)
- Monitors napari layer contrast_limits and visible events
- Preserves user settings when switching between wells
- Smart update detection (only regenerates when necessary)
Customizable Parameters:
- thumbnail_size: Default 80 pixels (speed/quality balance)
- max_threads: Worker count (default: 16)
- update_delay: Throttling delay (default: 100ms)
- handle_width: Splitter size (default: 8px)